Abstract
Background and objective: Epigenetic abnormality plays an important role in acute myeloid leukemia (AML) development. The lysine methyltransferase 2 (KMT2) family genes including KMT2A, KMT2B, KMT2C, KMT2D, SET1A and SET1B show significant impact on AML. KMT2A, also known as mixed-lineage leukemia (MLL) is the major player in MLL-fusion proteins found in AML, which are often associated with poor prognosis. Our group demonstrated KMT2C as a haplo-insufficient tumor suppressor gene in AML. KMT2D, a histone-3 lysine-4 methyltransferase, is one of the most frequently mutated genes like KMT2C in human cancers. Recent studies have established the essential tumor suppressor function of KMT2D in many solid tumors including lung adenocarcinoma and B cell lymphomas. However, the role of KMT2D in AML has so far not been completely elucidated. Thus, we sought to examine the role and molecular mechanisms of KMT2D in AML development.
Materials and methods: We firstly characterized the mutation and expression profiles of KMT2D in AML patients in cBioPortal. Then, we used shRNA interference technology and CRISPR/Cas9 genome editing to downregulate Kmt2d expression or disrupt Kmt2d in mouse hematopoietic stem and progenitor cells (HSPCs) respectively, and constructed mouse AML chimeric transplant models, exploring the biological function of KMT2D in AML development. Next, we performed RNA-seq to analyze the effects of Kmt2d expression on transcriptomes of mouse HSPCs and AML cells and combined RNA-seq with ATAC-seq and CUT-Tag data to discover downstream pathways and genes involved in Kmt2d deficiency-induced AML development. We conducted a series of relevant experiments in Kmt2d-deficient AML cells to verify the mechanisms. Lastly, we analyzed transcriptomes of AML patients with low KMT2D expression in TCGA-LAML cohort and phenotypes of KMT2D-mutated human AML cell line MOLM-13, evaluating whether the mechanisms of KMT2D in human AML were similar to these in mouse AML models.
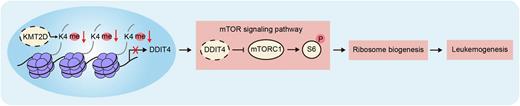
Results: There are often loss-of-function missense mutations of KMT2D and generally reduced expression of KMT2D in leukemia cells of AML patients compared with healthy control samples. AML patients with lower KMT2D expression levels were associated with poor prognosis. Compared to control recipient mice, mice transplanted with shKmt2d or sgKmt2d HSPCs developed AML significantly faster with shorter overall survival (shKmt2d-1: median 47 days after transplantation; p = 0.0004 and shKmt2d-2: median 74 days after transplantation; p = 0.0094; sgKmt2d-1: median 62 days after transplantation; p = 0.0027 and sgKmt2d-2: median 58 days after transplantation; p = 0.0027). Recipient mice developed AML as evidenced by phenotypic analyses of flow cytometry (CD117+; Gr-1/CD11b+; CD3ɛ/B220-) and pathological characteristics of peripheral blood, bone marrow, spleen and liver. Multi-omic analyses showed that Kmt2d deficiency,via histone methyltransferase activity, downregulated the expression of Ddit4, a negative regulator of the mTOR pathway, thus activated mTOR pathway and protein translation. We illustrated the downregulation of Ddit4 expression, activation of mTOR pathway, upregulation of ribosome biogenesis-related gene expression, enlargement of nucleolus and increase of protein synthesis rate in Kmt2d-dificient AML cells. MOLM-13 cells with KMT2D mutation and leukemia cells of AML patients with low KMT2D expression also had enhanced ribosome biogenesis.
Conclusions: Our study establishes the tumor suppressor function of KMT2D in AML, whose deficiency promotes AML development in mice.KMT2D can directly bind DDIT4, a negative regulator of mTOR signaling pathway, regulate gene expression, the activity of mTOR signaling pathway and ribosome biogenesis, and ultimately suppress AML development.
Disclosures
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal